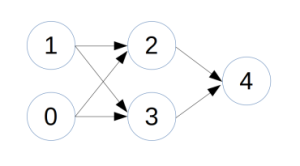
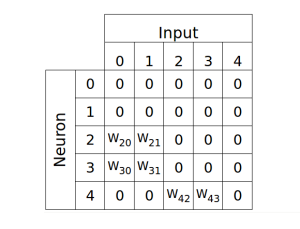
I’ve recently been working with neural networks and currently have a neural network class and a simple program to evolve the network weights.
The class and a few methods are defined in NeuralFunctions.py. The function createRandomWeights is currently set up to create a 9 neuron network when test = True and a completely random network when test = False.
The weights are defined as:
# Neural Network Class Functions
# NeuralFunctions.py
# Written in Python. See http://www.python.org/
# Placed in the public domain.
# Chad Bonner 1.31.2015
# Based on Back-Propagation Neural Networks by
# Neil Schemenauer <nas@arctrix.com>
print "NeuralNet v1.0"
import math
import random
import numpy as np
weightLimit = 5 #Somewhat arbitrary choice
#-------------Support Functions----------------
random.seed()
def createRandomWeights(n, weightLimit, test):
# n = number of neurons
# create matrix for weights
if test == True:
weights = np.array([[0.0 for i in range(n)] for j in range(n)])
weights[2,0] = random.uniform(-weightLimit,weightLimit)
weights[2,1] = random.uniform(-weightLimit,weightLimit)
weights[3,0] = random.uniform(-weightLimit,weightLimit)
weights[3,1] = random.uniform(-weightLimit,weightLimit)
weights[4,2] = random.uniform(-weightLimit,weightLimit)
weights[4,3] = random.uniform(-weightLimit,weightLimit)
else:
weights = np.array([[random.uniform(-weightLimit,weightLimit)
for i in range(n)]
for j in range(n)])
return weights
def createNets(numNets, sizeNet, numOut):
# Create population of neural nets
# Create a set of network connection weights with random weights
# Instantiate nets and load with weights
# Set test = True to use predefined weights
test = True
nets = [] #create empty variable to hold neural nets
for id in range(numNets):
#create weights for neuron connections
weights = createRandomWeights(sizeNet, weightLimit, test)
#instantiate a neural net
# id=0, number of neurons, number of outputs, dna
nets.append(NeuralNet(id, sizeNet, numOut, weights))
return nets
# our sigmoid function, tanh is a little nicer than the standard 1/(1+e^-x)
def sigmoid(x):
# init output variable
y = np.array([0.0]*len(x))
for i in range(len(x)):
y[i] = math.tanh(x[i])*2
return y
#------------ Neural Net Class Definition ------------------------------
class NeuralNet:
def __init__(self, id, n, no, weights):
# n = number of neurons
# no = number of outputs
self.id = id
self.n = n
self.no = no
self.weights = weights
# activations for nodes
self.ai = np.array([0]*self.n) #make all inputs neutral
self.an = np.array([0]*self.n) #neuron outputs
self.summ = np.array([0]*self.n) #init neuron activation levels to 0
def identifyNet(self):
print 'Net ', self.id
def update(self, neuronInputs):
# init neuron activation levels to 0
self.summ = np.array([0]*self.n)
# Apply inputs to input neurons
self.ai[:len(neuronInputs)] = neuronInputs
# Add neuron inputs to activation level
self.summ = self.ai + np.dot(self.weights, self.an)
# set neuron outputs
self.an = sigmoid(self.summ)
# net output is the output from the last no neurons
out = (self.an[self.n-self.no:])
#print "out", out
return out
# Evolution.py
#
# Written in Python. See http://www.python.org/
# Placed in the public domain.
# Chad Bonner 1.31.2015
from __future__ import division
import NeuralFunctions as nf
import weightedRandomChoice as wrc
import random
import numpy as np
import csv
import operator
from collections import defaultdict
# Setup
numGenerations = 50 #number of generations
numIterations = 4 #number of times to run each net
numNets =2000 #Size of the population
sizeNet = 5 #Number of neurons in each net
numOut = 1 #Number of outputs for each net
nets = [] #create empty variable to hold neural nets
nextGenNets = [] #create empty variable to hold neural nets
netFitness = {} #dictionary to hold net index and fitness
indexParents = [] #index of nets to be parents of next gen
parentDNA = [] #dna of parents
childDNA = [] #dna of children in the next gen
inputs = np.array([[0,0],[1,0],[0,1],[1,1]])
#============================================================================
#inputs = np.array([[0,0,0,0],[0,0,0,1],[0,0,1,0],[0,0,1,1],
# [0,1,0,0],[0,1,0,1],[0,1,1,0],[0,1,1,1],
# [1,0,0,0],[1,0,0,1],[1,0,1,0],[1,0,1,1],
# [1,1,0,0],[1,1,0,1],[1,1,1,0],[1,1,1,1]])
#============================================================================
def netEvaluate(nets, inputs):
#Apply input to each net and evaluate fitness
fitness = {} #dictionary to hold net index and fitness
for index, net in enumerate(nets):
#let signals progagate through net for iterations
#then look at output
fitness[index] = 0
for i in range(len(inputs)):
#print "Input:", i
for c in range(numIterations): #allow for propgation to output
out = net.update(inputs[i])
out = net.update(inputs[i])
#============================================================================ #Modify fitness here to suit application
#Arbitrary choice of matching for input pattern 0101. A correct match
#will have an output of 1 for an input of 0101 and an output of -1 for
#all other input patterns.
#Set out = [1,0] for even input
# if np.any(np.all(np.equal(inputs[i],
# [[0,0,0,0],[0,0,1,0],[0,1,0,0],[0,1,1,0],[1,0,0,0],
# [1,0,1,0],[1,1,0,0],[1,1,1,0]]), 1)):
# if out[0] >= 1 and out[1] < 0.2:
# fitness[index] = fitness[index] + 1 #award points
# else:
# if out[0] < 0.2 and out[1] >= 1:
# fitness[index] = fitness[index] + 1 #award
#============================================================================
#============================================================================
# #Modify fitness here to suit application
if np.any(np.all(np.equal(inputs[i], [[0,1],[1,0]]), 1)):
#print "Should be 1: Out =", out
if out >= [0.9]:
fitness[index] = fitness[index] + 1 #award points
#print "Rewarded for +1 output"
else:
#print "Should be 0: Out =", out
if out < [0.2]:
fitness[index] = fitness[index] + 1 #award
#print "Rewarded for 0 output"
if fitness[index] == 4:
fitness[index] = 15 #award a lot of points for correct answer
#============================================================================
return fitness
def writeFile(f, generation, fitness, fitIndex, nets, inputs, writeAll):
# Write data to CSV file
outputs = [] #create empty variable to hold net outputs
writer = csv.writer(f, delimiter=',')
if writeAll == True:
for net in nets:
writer.writerow(['Generation', 'Fitness', 'ID', 'SizeNet',
'NumOut', 'Weights'])
writer.writerow([generation, fitness, net.id, net.n, net.no])
for j in range(net.n):
writer.writerow(net.weights[j])
else:
writer.writerow(['Generation', 'Fitness', 'ID', 'SizeNet',
'NumOut', 'Weights'])
writer.writerow([generation, fitness, nets[fitIndex].id,
nets[fitIndex].n,
nets[fitIndex].no])
for i in range(len(inputs)):
for c in range(numIterations):
nets[fitIndex].update(inputs[i])
outputs.append(nets[fitIndex].update(inputs[i]))
writer.writerow(outputs)
print outputs
for j in range(nets[fitIndex].n):
writer.writerow(nets[fitIndex].weights[j])
return
def breed(netFitness, nets):
# Create new population by combining
# parents's dna (sexual reproduction).
# Each net can combine with any other net, even itself.
#print netFitness
pairs = zip(netFitness.values(),netFitness.keys())
pairs.sort()
pairs.reverse()
nextGenNets = [] # clear next gen variable for use
for c, net in enumerate(nets):
mom = wrc.weighted_random_choice(pairs)
#print "mom", mom
dad = wrc.weighted_random_choice(pairs)
#print "dad", dad
childDNA = np.array([[0.0 for i in range(net.n)]
for j in range(net.n)])
for neuron in range(net.n):
#take one neuron from each parent and combine to form child
childDNA[neuron,:] = nets[random.choice((mom,dad))].weights[neuron,:]
# now create new net using child dna
# instantiate a neural net
nextGenNets.append(nf.NeuralNet(c, net.n, net.no, childDNA))
return nextGenNets
# ------------------------- Main Routine ------------------------------
#def main():
#Open file to save data
csvfile = open('data_out.csv', 'wb')
#Create and evaluate initial population of neural nets
print "Genesis!"
nets = nf.createNets(numNets, sizeNet, numOut)
# Apply input to each net and evaluate fitness
netFitness = netEvaluate(nets, inputs)
fitHistogram = defaultdict(int) #variable to hold count of fitness values
#Count frequency of fitness values
for value in netFitness.values(): fitHistogram[value] += 1
#print fitHistogram
print "Generation Fitness:", "%.3f"%((sum(netFitness.values())/numNets))
# Identify net with maximum fitness and save to log file
mostFit = max(netFitness.iteritems(), key=operator.itemgetter(1))[0]
mostFitness = netFitness[mostFit]
print "Most Fit:", mostFit
writeFile(csvfile, 0, mostFitness, mostFit, nets, inputs, False)
# Go forth and multiply
for generation in range(1, numGenerations):
#Breed next generation
nets = breed(netFitness, nets)
print " "
print "Generation:", generation, "born"
# Apply input to each net and evaluate fitness
netFitness = netEvaluate(nets, inputs)
fitHistogram = defaultdict(int) #variable to hold count of fitness values
#Count frequency of fitness values
for value in netFitness.values(): fitHistogram[value] += 1
print fitHistogram
print "Generation Fitness:", "%.3f"%((sum(netFitness.values())/numNets))
# Identify net with maximum fitness and save to log file
mostFit = max(netFitness.iteritems(), key=operator.itemgetter(1))[0]
mostFitness = netFitness[mostFit]
print "Most Fit:", mostFit
writeFile(csvfile, generation, mostFitness, mostFit, nets, inputs, False)
#print "Best net saved"
#print "Iteration", generation, "complete!"
csvfile.close()
print "Done!"
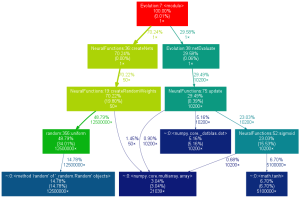
Currently it takes about 12 seconds to run a population of 50 nets of 500 neurons each for 50 cycles. I’m using numpy to speed things up. I’m sure there are still opportunities for improvement. Creating the nets actually takes longer than running them as shown below (click to enlarge).